We’ve just released version 0.4 of mldr. Here’s a brief look at the changes. Don’t forget to update!
Reimplementation of evaluation metrics
The set of evaluation metrics included in mldr has been accumulating issues and bug reports in the last months. This fact has encouraged us to fully revise and reimplement these metrics. The new implementations have been thoroughly tested and compared to other in different libraries.
Evaluation metrics are now exported individually, but you can keep using the mldr_evaluate function for a full report on the performance of a classifier. Additionally, the roc method has been implemented for the "mldr" class, and it allows to obtain the ROC curve for a dataset and its predictions (provided the pROC package is installed).
Treatment of undefined values in metrics
Several performance metrics can sometimes lead to undefined results, usually due to potential divisions by zero. With the aim of facilitating experimentation with different classifiers among diverse platforms, we have provided parameters that customize the behavior of performance metrics in these cases. In particular, mldr mimics the behavior of MULAN metrics by default, but other options are available. Let’s look at some examples:
true_labels <- matrix(c(
1,1,1,
0,0,0,
1,0,0,
1,1,1,
0,0,0,
1,0,0
), ncol = 3, byrow = TRUE)
predicted_labels <- matrix(c(
1,1,1,
0,0,0,
1,0,0,
1,1,0,
1,0,0,
0,1,0
), ncol = 3, byrow = TRUE)
# strategies for undefined values: "diagnose", "ignore", "na"
precision(true_labels, predicted_labels, undefined_value = "diagnose")
# single value to replace undefined values: e.g. 0, 1
recall(true_labels, predicted_labels, undefined_value = 0)
# custom strategy for undefined values
fmeasure(
true_labels, predicted_labels,
undefined_value = function(tp, fp, tn, fn) as.numeric(fp == 0 && fn == 0)
)
In the first example, we are using one of mldr’s built-in strategies to treat undefined values. The "diagnose" strategy is the default behavior, and assigns undefined values a replacing value of 1 or 0 according to the accuracy of the prediction (whether no labels were predicted for an instance with no relevant labels). On the contrary, "ignore" would not count those cases for the averaging process (which is different from assigning them value zero and counting them). Last, the "na" strategy will propagate NA if encountered.
The second example is simple, it replaces any NA value with the specified number. A reasonable value can be 0, but this may wrongly penalize the classifier in datasets with very sparse labels.
The third example is the custom approach: you can provide a function accepting 4 integers (TP, FP, TN, FN), which will be called when an undefined value is encountered, with the true positives, false positive, true negatives and false negatives respectively. It should generally return a numeric value (in the range [0, 1] in order for the mean to make sense).
Improvements on read and write of ARFF files
The parser for ARFF files is now more robust, including support for single-quoted and double-quoted attributes, as well as a negative amount of labels in the MEKA header (indicating labels are at the end of the attribute list). Additionally, the user can now choose to read categorical attributes as factors (passing the stringsAsFactors parameter).
Exporting to ARFF has seen some improvements as well, but you may want to check out mldr.datasets, which is able to export to a variety of other formats and provides more options.
Until now, multilabel datasets have been provided in different file formats for different pieces of software. mldr was created with compatibility in mind and allowed to read two widely-known formats: datasets from Mulan and MEKA repositories in ARFF format.
With the creation of the Ultimate Multilabel Dataset Repository (RUMDR) and a new R package, mldr.datasets, a huge set of multilabel datasets are now available in a common format and with the possibility of being converted into many more.
mldr.datasets
Multilabel datasets
install.packages("mldr.datasets")
library(mldr.datasets)
Note: mldr.datasets does not depend on mldr, but it’s useful to have both of them installed to access all functionality.
After installing and loading the package, some pre-loaded datasets will be available directly in the environment:
- birds
- cal500
- emotions
- flags
- genbase
- langlog
- medical
- ng20
- slashdot
- stackex_chess
These are accessible via their names and the usual members of "mldr" objects ($measures, $labels, $datasets…). Additionally, a toBibtex() method is provided for fast access to the citation information for each dataset.
Larger datasets are available to download from the repository (you can consult the complete list of datasets or call mldrs()) via the check_n_load.mldr() function.
Partitioning functions
The random.kfolds() and stratified.kfolds() functions partition multilabel datasets following a random strategy and a stratified one, respectively.
Exporting functions
The write.mldr() function is able to export "mldr" and "mldr.folds" objects into several file formats: Mulan, MEKA, KEEL, LibSVM and CSV. This way, regular, partitioned and preprocessed datasets can be saved for later use in any well-known multilabel classification tool.
mldr 0.3.18
We’ve updated mldr to integrate functionality from mldr.datasets when it’s installed. Thus, now calling mldr() with simply a dataset name will trigger a search within the datasets in the repository. If a dataset isn’t found, the function will attempt to read the dataset locally (this behavior can be forced using the force_read_from_file parameter).
Other changes in this update include exposing the read.arff() function, able to read ARFF files and differentiate input and output features without calculating any related measure, as suggested in issue 26; several fixes related to dataset reading and calculation of measures, and slight changes in some calculations. For detailed information visit our changelog or the commit history.
We have released a new version of mldr, 0.2.51, already live on CRAN. It fixes a recently found bug and adds functionality to the plotting function.
Plot improvements
Multiple plots in one call
The plot function now allows a vector of plot types as its type parameter. This results in the generation of multiple plots, with a pause between them if needed to display all of them separately. The following is an example of this functionality:
plot(emotions, type = c("LB", "LSB", "LH", "LSH"))
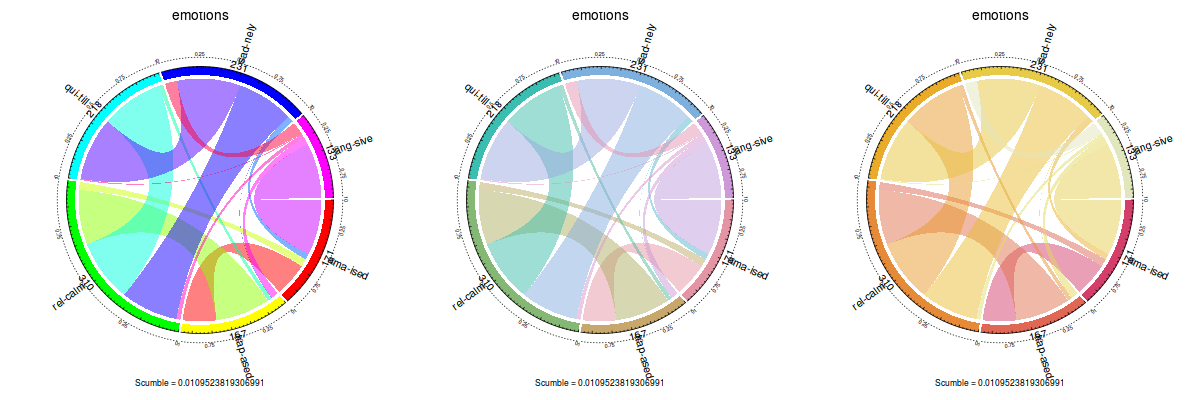
Coloring plots
Until now, color in plots were fixed and couldn’t be changed by the user. The update adds the col and color.function parameters. The former can be used on all plot types except for the label concurrence plot, and must be a vector of colors. The latter is only used on the label concurrence plot and accepts a coloring function, such as rainbow or heat.colors, or the ones provided by the colorspace package:
layout(matrix(c(1, 2, 3), 1, 3))
plot(emotions, color.function = rainbow)
plot(emotions, color.function = colorspace::rainbow_hcl)
plot(emotions, color.function = colorspace::heat_hcl)
Bug fixes
A bug was found when loading sparse datasets with a certain formatting. This has been fixed on the update and shouldn’t be a problem anymore.
A small update has been released on CRAN, including a GUI redesign and better ability to read datasets from it.
GUI design changes
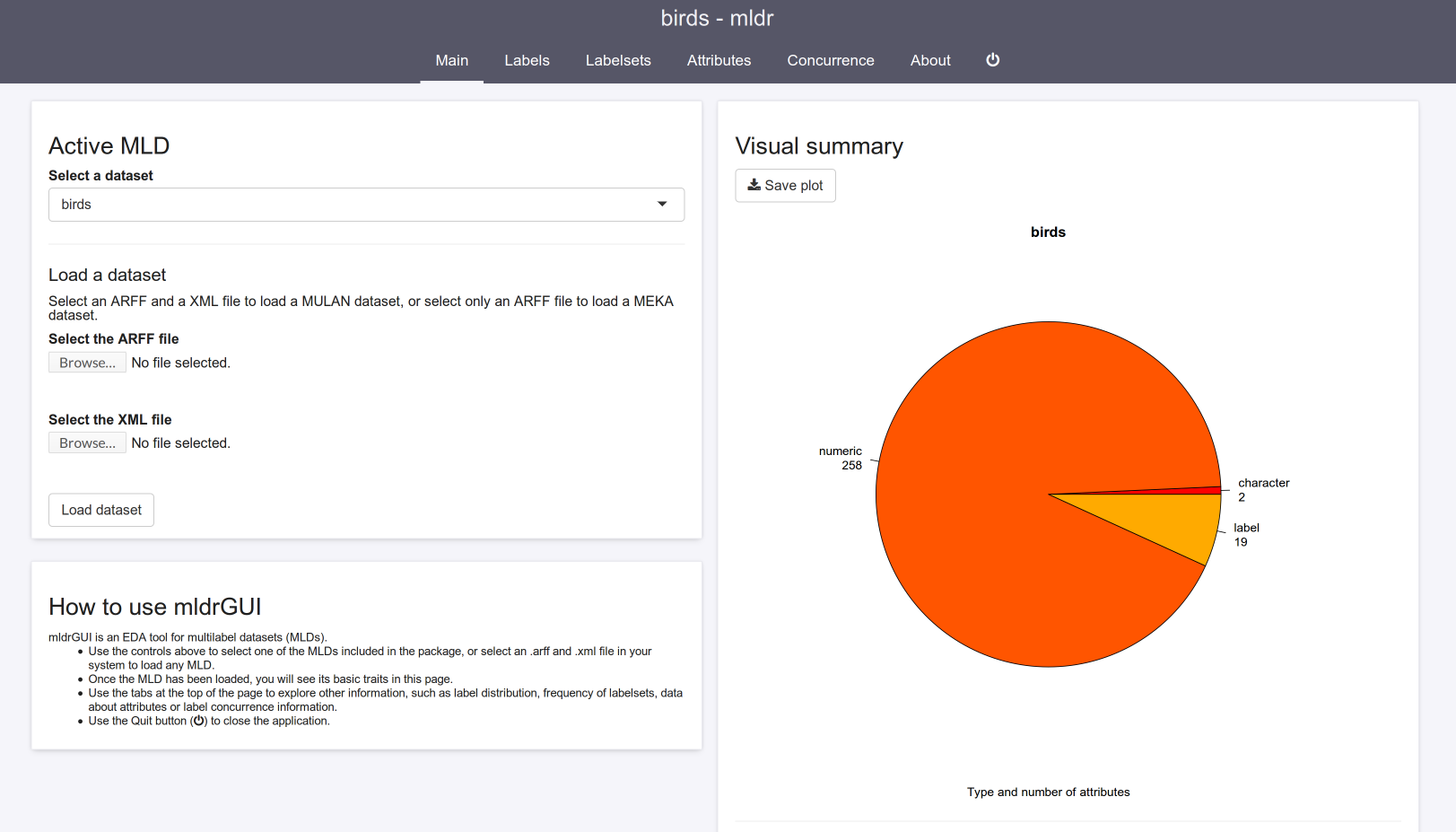
Some changes have been made to the web interface of mldr. This design saves more space, better highlights content and is less distracting while being visually appealing.
Loading MEKA datasets from GUI
Users are now able to load MEKA-style datasets (in ARFF format without a XML file for labels) from the web interface as well. To do this, just upload the ARFF file and click Load dataset without selecting any XML file.
A new version of mldr was just released and is now live on CRAN. This update adds new functions able to assess multilabel classifier performance, allows to create mld objects out of ARFF files with different structures (see example below), and fixes several bugs.
mldr now includes the mldr_evaluate function, which analyzes the performance of classifier predictions via several well-known measures (Accuracy, Precision, Recall, F-measure, Hamming Loss among others). Using it is simple: just call it with the test dataset and the predictions generated by the classifier. The function will return a list with all 20 measures identified by their names.
res <- mldr_evaluate(emotions, predictions)
New parameters to identify labels
Labels in ARFF files can be structured in several ways, so now the mldr constructor allows the use of three new parameters that will ease the read of a multilabel dataset: label_indices, label_names and label_amount. The first one enables the user to specify exactly the indices the labels will be taking in the dataset; the second one identifies labels by using their names, and the last one takes a number of labels to be read from the last attributes in the ARFF file.
corel5k <- mldr("corel5k", label_amount = 374)
emotions <- mldr("emotions", label_indices = c(73, 74, 75, 76, 77, 78))
New vignette: Working with Multilabel Datasets in R
A vignette has been added to mldr as well. The document instructs the reader on how the package works and provides examples to ease the learning. It can be loaded with the command vignette("mldr") or downloaded from CRAN.

